Dickinson Lab
From the Dickinson Lab @ UT Austin
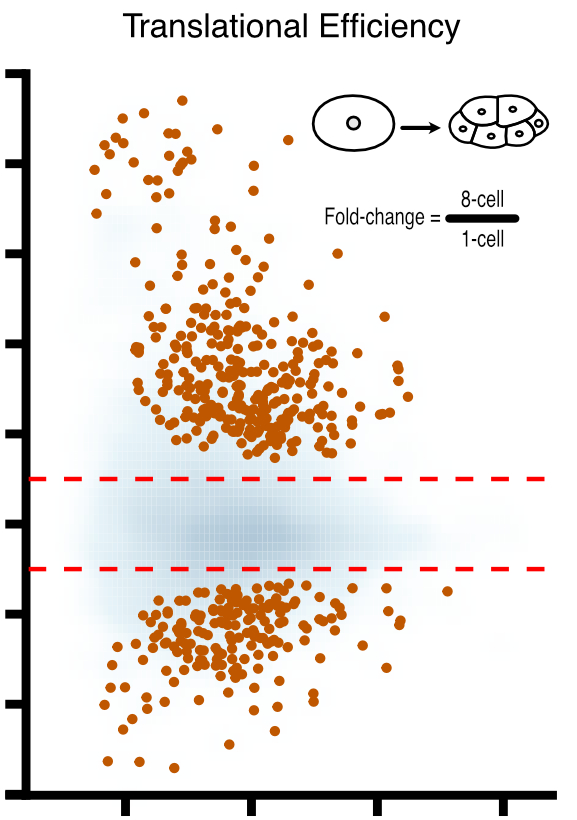
Landscape and regulation of new protein translation in the early C. elegans embryo
Shukla Y, Ghatpande V, Hu CF, Dickinson DJ, Cenik C. bioRxiv 2024
Animal embryos are transcriptionally quiescent and rely on maternal mRNAs and proteins to develop. C. elegans embryos are especially interesting in this regard because they irreversibly specify cell fates before activating their own genomes. To help understand how this can happen, we teamed up with Can Cenik’s lab to measure new protein translation in early embryos. The results reveal how maternal mRNAs are regulated by RNA binding proteins during early development, and provide a valuable resource for others in the field.
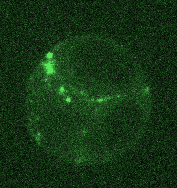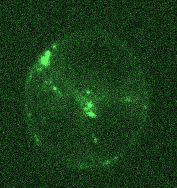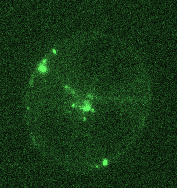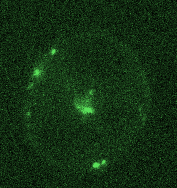
Opposing actomyosin pools generate cortical flows that establish epithelial polarity
Shi Y, Ohler LS, de Jesus BN, Dickinson DJ. bioRxiv 2024
Our first biological study using mouse embryonic stem cells! We used live imaging of 3D cultures to reveal how actomyosin contractility remodels cell-cell junctions during epithelial polarization. We found that there are two pools of active myosin in these cells: An apical pool that depends on myosin light chain kinase, and a basal pool that depends on Rho kinase. We think that different contractility of these two myosin pools leads to transport or zippering of tight junctions to establish the apical cell-cell junction.

Temporally distinct roles of Aurora A in polarization of the C. elegans zygote
Manzi NI, de Jesus B, Shi Y, Dickinson DJ. Development 2024
The cell cycle kinase Aurora A is implicated in symmetry breaking (that is, initiation of polarity establishment). However, different groups reported different phenotypes following Aurora A RNAi, ranging from no posterior domain (no symmetry breaking) to two posterior domains (excess symmetry breaking). We used chemical inhibitors and auxin-inducible degradation to show that acute inhibition of Aurora A prevents symmetry breaking, while chronic Aurora A depletion in the germline leads to excess symmetry breaking. Thus, Aurora A has distinct functions (and probably distinct mechanisms of action) in the germline and in embryos.
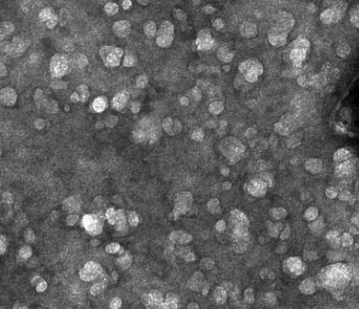
Membrane extraction in native lipid nanodiscs reveals dynamic regulation of Cdc42 complexes during cell polarization
Deutz LN, Sarıkaya S, Dickinson DJ. Biophysical Journal 2023
Many critical cell signaling events happen at the plasma membrane. Studying the protein complexes that drive those events presents a major technical challenge, because conventional biochemical approaches are ill-suited to studying proteins in intact membranes. We combined polymer membrane scaffolds with laser-induced cell lysis to create cell-derived, native lipid nanodiscs, which we could then study using single-molecule biochemistry. This allowed us to study the Cdc42/Par6 complex, which was previously undetectable, and revealed that this complex is developmentally regulated.

Apical PAR protein caps orient the mitotic spindle in C. elegans early embryos
Stolpner NJ, Manzi NI, Su T, Dickinson DJ. Current Biology 2023
Most if not all animal embryos establish inside-outside polarity at an early stage. In C. elegans, inside-outside polarity is established at the 8-cell stage, but its function was unknown. We found that apicobasal polarity is required for proper mitotic spindle positioning in 8-cell embryos. Our results show that polarity in this stage is established via a mechanism that is very different from the 1-cell embryo, which polarized just a few minutes earlier, raising the question of how polarity pathways are rapidly remodeled during development.
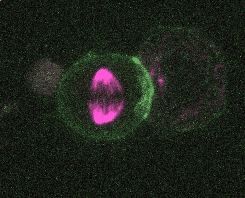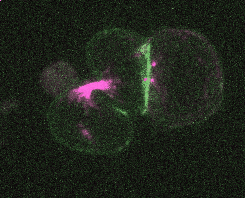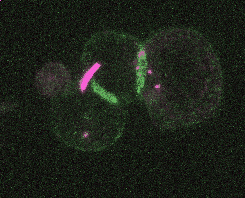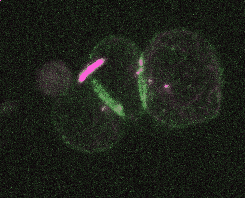
Efficient and rapid fluorescent protein knock-in with universal donors in mouse embryonic stem cells
Shi Y, Kopparapu N, Ohler L, Dickinson DJ. Development 2023
Endogenous gene tagging has been a cornerstone of our experimental toolkit since 2013, when Dan developed the first fluorescent protein knock-in protocol for C. elegans. A decade later, we’ve extended this strategy to a new model system: mouse embryonic stem cells (mESCs). Although the techniques required for gene tagging in mESCs are different from C. elegans, the outcome is the same: We can perform live imaging of endogenous proteins – and coming soon, single-cell biochemistry! – without overexpression artifacts.
Non-invasive chimeric HaloTag labeling to study clustering and diffusion of membrane proteins
Chang Y, Dickinson DJ. STAR Protocols 2022
In our Cell Reports paper published earlier in 2022, we showed how we could use dual-colored labeling of HaloTag to simultaneously estimate the sizes of PAR-3 clusters and track their diffusion on the cortex. Here we provided a protocol for this dual-labeling strategy, which has potential applications for many proteins beyond PAR-3.
A particle size threshold governs diffusion and segregation of PAR-3 during cell polarization
Chang Y, Dickinson DJ. Cell Reports 2022
PAR-3 oligomers are essential for C. elegans zygote polarization, but since oligomers have a range of sizes, it was difficult to determine at the biophysical level how different-sized oligomers contribute to polarity. Ivy addressed this challenge by engineering PAR-3 oligomers of defined sizes and quantitatively analyzing their diffusion. She found that oligomers as small as a trimer undergo directed motion due to cortical flow and can support cell polarization. She also verified this by developing an innovative dual-labeling strategy that allows simultaneous tracking and size measurement of endogenous PAR-3.
Rapid extraction and kinetic analysis of protein complexes from single cells
Sarıkaya S, Dickinson DJ. Biophysical Journal 2021
Our 2017 method for single-cell biochemistry was a powerful approach for measuring protein-protein interactions in vivo. But an important limitation was that the time from (manual) cell lysis to data collection was 3-5 minutes – too slow for protein complexes that have weaker affinity. We found that we could use a pulsed infrared laser to lyse cells and then begin collecting data immediately, allowing us to measure the kinetics of protein complex dissociation. Laser lysis also improves reproducibility and allows us to document the stage at which each cell or embryo is lysed.
Single-cell single-molecule pull down (sc-SiMPull) for detection of protein complexes from embryonic lysates
Stolpner N, Dickinson DJ. Methods in Mol. Biol. 2022
Presents a protocol for performing sc-SiMPull using mechanical lysis of C. elegans embryos, as first developed in Dan’s 2017 Developmental Cell paper. See later papers from our group for updates and improvements.
Electron microscopy snapshots of single particles from single cells
Yi X, Verbeke EJ, Chang Y, Dickinson DJ*, Taylor DW*. J. Biol. Chem. 2019
Reports that single-embryo lysates (prepared using our microfluidic lysis chips) contain sufficient material to observe 3D structures of native protein complexes. We visualized ribosomes and proteasomes from single-embryo lysates as a proof of principle.
From our “Glow Worms” Freshman Research stream
Using CRISPR knock-in of fluorescent tags to examine is-form-specific expression of EGL-19 in C. elegans
McDonald K, Larkin K, Dickinson DJ, Golden A, Bai X, Doonan, R. (2023)
microPublication Biology 10.17912/micropub.biology.000858.
As part of a collaboration with Andy Golden’s lab at NIH, our team generated knock-in strains to specifically label different isoforms of EGL-19, allowing the Golden lab to determine where each isoform is expressed within the nervous system.
mScarlet and split fluorophore mScarlet resources for plasmid-based CRISPR/Cas9 knock-in in C. elegans
Witten G, DeMott E, Huang G, Zelasko F, de Jesus B, Mulchand C, Schuck L, Pullman S, Perez A, Mahableshwarkar P, Wu Z, Cardona EA, Pierce JT, Dickinson DJ, Doonan R. (2023)
microPublication Biology 10.17912/micropub.biology.000871.
Our team of undergraduate researchers generated and tested a plasmid toolkit for inserting the bright red fluorescent protein mScarlet-I into the C. elegans genome.
Highly improved cloning efficiency for plasmid-based CRISPR knock-in in C. elegans
DeMott E, Dickinson DJ, Doonan, R. (2021)
microPublication Biology 10.17912/micropub.biology.000499.
Cloning is often a rate-limiting step in molecular biology research, and genome editing is no exception. Our team extensively optimized the DNA concentrations and conditions for assembling CRISPR/Cas9 targeting vectors and repair templates for C. elegans, resulting in nearly 100% efficiency.
Improved CRISPR/Cas9 knock-in efficiency via the self-excising cassette (SEC) selection method in C. elegans
Huang G, de Jesus B, Koh A, Blanco S, Rettmann A, DeMott E, Sylvester M, Ren C, Meng C, Waterland S, Rhodes A, Alicea P, Flynn A, Dickinson DJ, Doonan, R. (2021)
microPublication Biology 10.17912/micropub.biology.000460.
The Self-excising cassette is a selectable marker, invented by Dan in 2015, that enables streamlined and robust generation of genome-edited C. elegans strains. Our team of freshman researchers, led by Assistant Professor of Practice and staff scientist Ryan Doonan, has extensively optimized this protocol and achieved a more than 10-fold increase in efficiency compared to our original procedure.
Work from others that we’ve contributed to
An expanded auxin-inducible degron toolkit for Caenorhabditis elegans
Ashley GE, Duong T, Levenson MT, Martinez MAQ, Johnson LC, Hibshman JD, Saeger HN, Palmisano NJ, Doonan R, Martinez-Mendez R, Davidson BR, Zhang W, Ragle JM, Medwig-Kinney TN, Sirota SS, Goldstein B, Matus DQ, Dickinson DJ, Reiner DJ, Ward JD. (2021)
Genetics 217(3): iyab006.
The Auxin-Inducible Degron (AID) system is emerging as a powerful tool for controlled depletion of proteins of interest. This system requires expressing a ubiquitin ligase, TIR1, that derives from plants. We created a TIR1 expression construct that contains a built-in BFP::AID reporter, allowing TIR1 expression and activity to be monitored without using the GFP or RFP imaging channels. The lab was brand new at the time and we didn’t have the bandwidth to further develop this ourselves. We shared the constructs and strategy with the Ward and Reiner labs, who developed a really nice toolkit based on our idea. Kudos to their groups for building an awesome resource.
Ras-dependent cell fate decisions are reinforced by the RAP-1 small GTPase in Caenorhabditis elegans
Rasmussen NR, Dickinson DJ, Reiner DJ. (2018)
Genetics 210(4): 1339-54
From Dan’s Postdoc
Goldstein Lab @ UNC Chapel Hill
Optogenetic dissection of mitotic spindle positioning in vivo
Fielmich LE, Schmidt R, Dickinson DJ, Goldstein B, Akhmanova A & van den Heuvel S. (2018). eLife e38198.
Pubmed Full Text
One of my side projects as a postdoc was devising an algorithm to design protein-coding sequences that would evade silencing in the C. elegans. I contributed this algorithm to an optogenetics project in the van den Heuvel lab; it was used to design sequences that allowed expression of LOV domain-based optogenetic tools in C. elegans.
SapTrap assembly of repair templates for Cas9-triggered homologous recombination with a self-excising cassette
Dickinson DJ, Slabodnick MM, Chen AH, Goldstein B. (2018). microPublication Biology 10.17912/W2KTON
Pubmed Full Text
Light-dependent cytoplasmic recruitment enhances the dynamic range of a nuclear import photoswitch
Yumerefendi H, Wang H, Dickinson DJ, Lerner AM, Malkus P et al. (2018). Chembiochem 19(12): 1319-25.
Pubmed Full Text
A CRISPR tagging-based screen reveals localized players in Wnt-directed asymmetric cell division
Heppert JK, Pani AM, Roberts AM, Dickinson DJ, Goldstein B. (2018). Genetics 208(3): 1147-64.
Pubmed Full Text

A single-cell biochemistry approach reveals PAR complex dynamics during cell polarization
Dickinson DJ*, Schwager F, Pintard L, Gotta M, Goldstein B. (2017). Developmental Cell 42(4): 416-34.
Pubmed Full Text
Reports an approach for analyzing protein-protein interactions in single, staged C. elegans zygotes. Cells are lysed in nanoliter volumes using microfluidics, and contents analyzed using single-molecule pull-down. This approach was used to discover that oligomerization of the PAR-3 protein is surprisingly dynamic: it is upregulated specifically during polarity establishment. PAR-3 oligomerization is essential for proper polarity establishment and is linked to the cell cycle via direct phosphorylation of PAR-3 by PLK-1.

Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system
Heppert JK, Dickinson DJ, Pani AM, Higgins CD, Steward A. et al. (2016). Molecular Biology of the Cell 27(22): 3385-94.
Pubmed Full Text
Quantitative head-to-head comparison of our favorite green and red fluorescent proteins for imaging C. elegans embryos.
MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation
Marston DJ, Higgins CD, Peters KA, Cupp TD, Dickinson DJ et al. (2016). Current Biology 26(16): 2079-89.
Pubmed Full Text
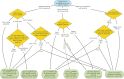
CRISPR-based methods for genome engineering
Dickinson DJ, Goldstein B. (2016). Genetics 202(3): 885-901.
Pubmed Full Text
Comprehensive review of CRISPR methods for C. elegans, including (still current!) recommendations for which strategies are most appropriate for which kinds of modifications.
Identifying regulators of morphogenesis common to vertebrate neural tube closure and Caenorhabditis elegans gastrulation
Sullivan-Brown J, Tandon P, Bird KE, Dickinson DJ, Heppert JK et al. (2016) Genetics 202(1): 123-39.
Pubmed Full Text
Crescerin uses a TOG domain array to regulate microtubules in the primary cilium
Das A, Dickinson DJ, Wood C, Goldstein B, Slep KC. (2015). Molecular Biology of the Cell 26(23): 4248-64.
Pubmed Full Text
Results of a collaboration between Dan and Alakanada Das, who was a graduate student in Kevin Slep’s lab at the time. Alka solved the crystal structure of a key microtubule-binding domain in the Crescerin protein. Dan helped her learn how to work with worms, and designed and constructed some targeted mutants in the worm Crescerin gene che-12 to test specific hypotheses about its function.
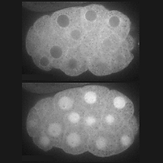
Control of protein activity and cell fate specification via light-mediated nuclear translocation
Yumerefendi H‡, Dickinson DJ‡, Wang H, Zimmerman SP, Bear JE et al. (2015). PLoS One 10(6): e0128443
‡Equal Contribution
Pubmed Full Text
Results of a collaboration between Dan and Hayretin Yumerefendi, a postdoc in Brian Kuhlman’s lab. Hayretin engineered a small protein tag that localizes to the cytoplasm in the dark but translocates reversibly to the nucleus on stimulation with blue light. Dan designed and carried experiments to test whether this switch can be used to control transcription factor activity in vivo (spoiler alert – it can).

Streamlined genome engineering with a self-excising drug selection cassette
Dickinson DJ*, Pani AM, Heppert JK, Higgins CD, Goldstein B. (2015). Genetics 200(4): 1035-49.
Pubmed Full Text
Reports a strategy for modifying the C. elegans genome, which was designed and optimized to minimize hands-on labor. The key innovation was a new self-excising cassette (SEC) for drug selection. The SEC works in a wild-type background, produces a visible phenotype to facilitate mutant isolation, and can be removed after use by simply heat shocking the worms. The SEC strategy is particularly effective for fluorescent protein tagging, but can also be used to produce point mutations.
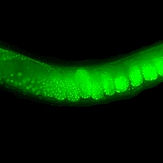
Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination.
Dickinson DJ*, Ward JD, Reiner DJ, Goldstein B. (2013). Nature Methods 10: 1028-34.
Pubmed Full Text
Reports that the CRISPR/Cas9 system can be used to trigger homologous recombination in C. elegans. By supplying an appropriate homologous repair template, a variety of designer mutations were produced, including endogenous GFP fusions and targeted phosphorylation site mutations. This was one of the first papers reporting that CRISPR could be used in C. elegans.
From Dan’s Ph.D.
Nelson and Weis labs @ Stanford
Measuring Protein Binding to F-actin by Co-sedimentation.
Heier JA, Dickinson DJ, Kwiatkowski AV. (2017). Journal of Visualized Experiments. (123). doi: 10.3791/55613.
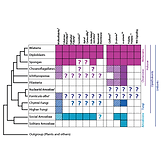
Dickinson DJ*, Nelson WJ, Weis WI. (2012). Bioessays 34: 833-40.
Presents a new hypothesis for the evolution of metazoan multicellularity from unicellular ancestors. Some modern organisms, including Dictyostelium and some fungi, are “facultative” multicellular organisms – that is, they become multicellular only under certain conditions. Based on similarities between epithelial tissues in Dicty and animals, it is proposed that this facultative mode of multicellularity may be ancestral.

Dickinson DJ, Robinson DN, Nelson WJ, Weis WI. (2012). Developmental Cell 23: 533-46.
Identifies a link between alpha-catenin, IQGAP and myosin during multicellular development of Dictyostelium. Alpha-catenin recruits IQGAP to the basolateral membrane of Dicty epithelial cells, and IQGAP in turn acts to exclude myosin from the basolateral cortex. This results in apical actomyosin enrichment, which controls the diameter of the epithelial tube and thereby controls the shape of the entire fruiting body.
Protein evolution in cell and tissue development: Going beyond sequence and transcriptional analysis.
Dickinson DJ, Weis WI, Nelson WJ. (2011). Developmental Cell 21: 32-34.
A philosophical essay that argues for a more experimental approach towards understanding protein evolution that considers protein function in the context of a cell. Read carefully and you’ll see some early traces of the ideas that fueled Dan’s postdoctoral work.
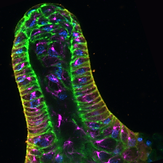
A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins..
Dickinson DJ, Nelson WJ, Weis WI. (2011). Science 331: 1336-39.
Reports the identification of a polarized epithelial tissue during multicellular development of the social amoeba Dictyostelium. Epithelial cells are organized into a tube at the tip of the developing fruiting body, where they secrete cellulose and ECM proteins that surround the stalk. A newly-identified alpha-catenin homolog, together with a previously-identified beta-catenin homolog, are essential for normal epithelial organization and polarity, suggesting that aspect of animal multicellularity may have been present in the common ancestor of Dicty and animals.